PhD project -
RBC deformation in microchips
Despite the extensive research conducted over the past five decades to investigate the mechanical characteristics of Red Blood Cells (RBCs), the existing diagnostic landscape lacks the capability to offer comprehensive insights into individual cell behaviour. Therefore, the development of new diagnostic tools able to perform analysis at the scale of a single cell is urgently needed. When a RBC suffers from a disorder, the mechanical properties can change and trigger fatal consequences in our bodies. In order to investigate the mechanical response of the RBCs, in this project, RBCs are simulated in a Poiseuille flow through a microfluidic constriction. These simulations aim to compute the flow and deformation of synthetic discretized cells using Computational Fluid Dynamics (CFD) software, which in this case is OpenFoam, coupling via the Immersed Boundary Method (IBM).
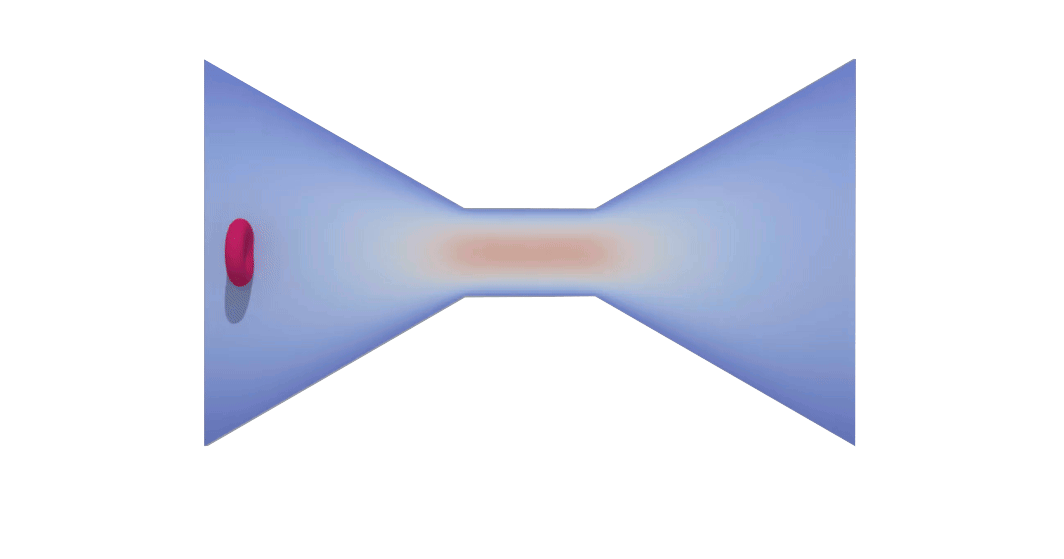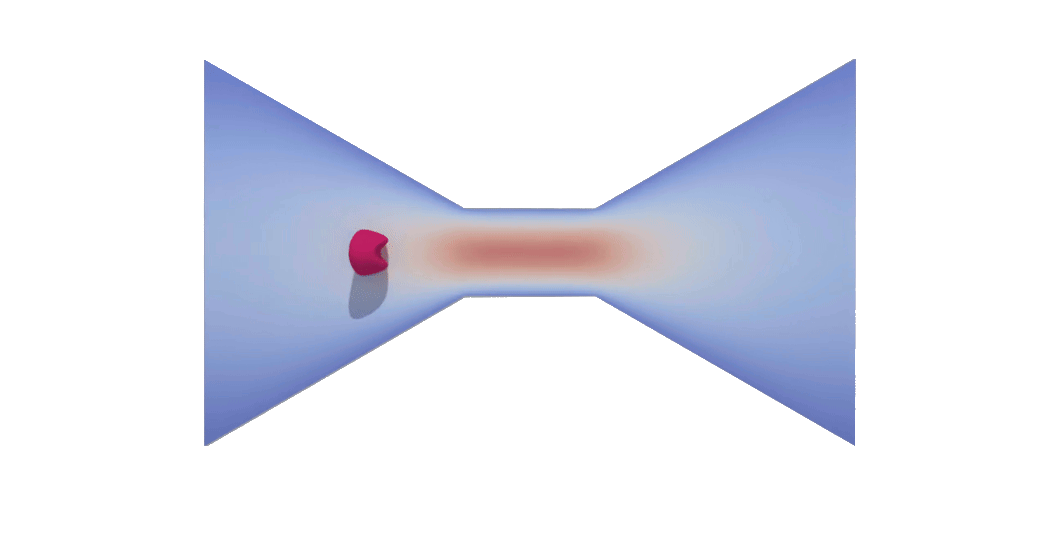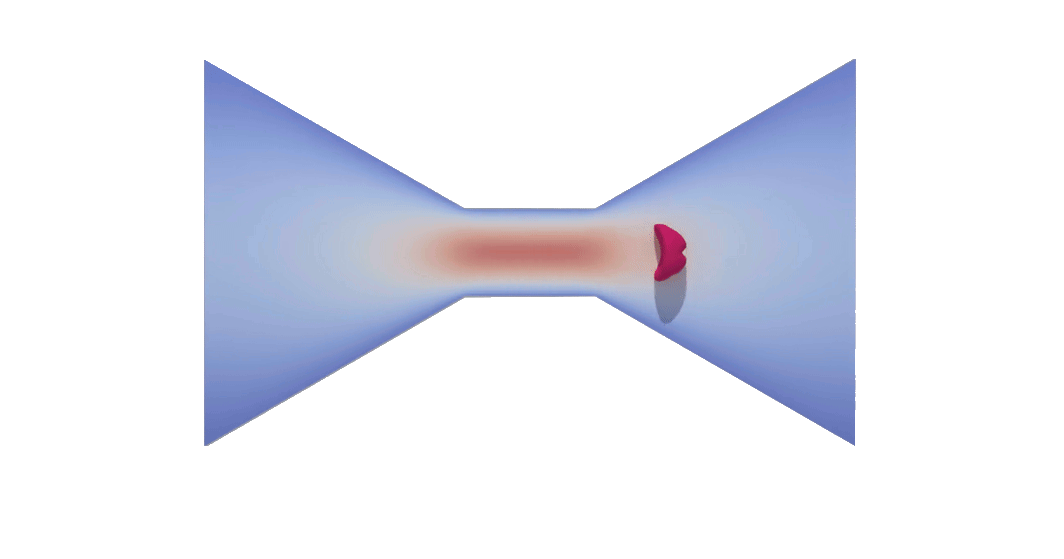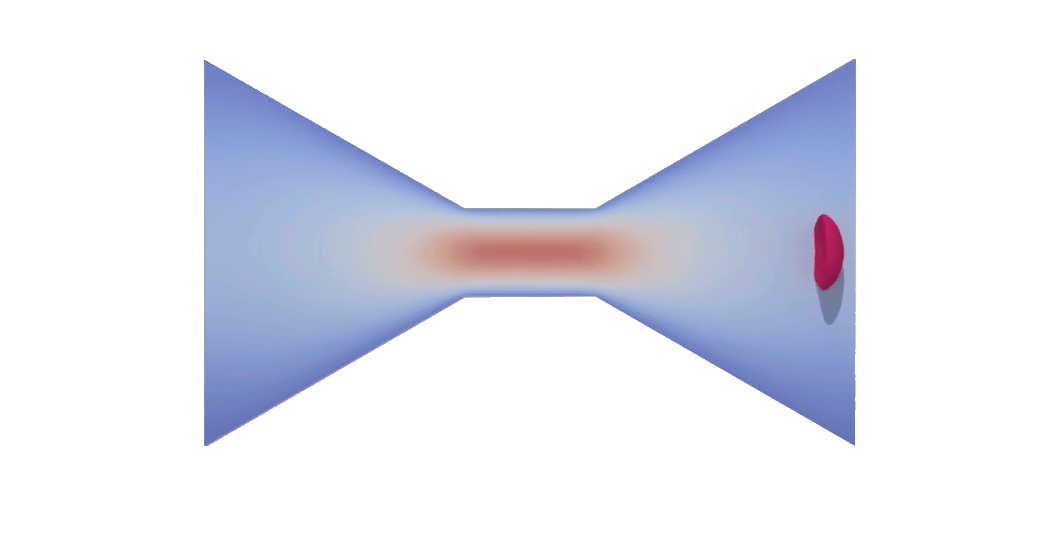
RBC flowing through a microchip constriction. The color gradient shows the velocity inside the microchip.
The velocity in the constriction section is higher (more red).
The RBCs were implemented in Mpcats, which is based on Python, and the fluid is solved with the simpleFoam solver of OpenFoam. The coupling handles the force from the cells and the velocity from the fluid to build up a bridge between CFD and cells. Via Lagrangian (Mpacts)-Eulerian (CFD) description, the simulation is able to solve the Navier-Stokes (N-S) equations, update the velocity field, transfer this to the Lagrangian points, interpolate the new positions of the MPACTS nodes, calculate the forces from the nodes, couple back this forces to the Eulerian fluid and use them in the next timestep to solve N-S equations again. This configuration enables the implementation of the interaction between multiple cells and the fluid:
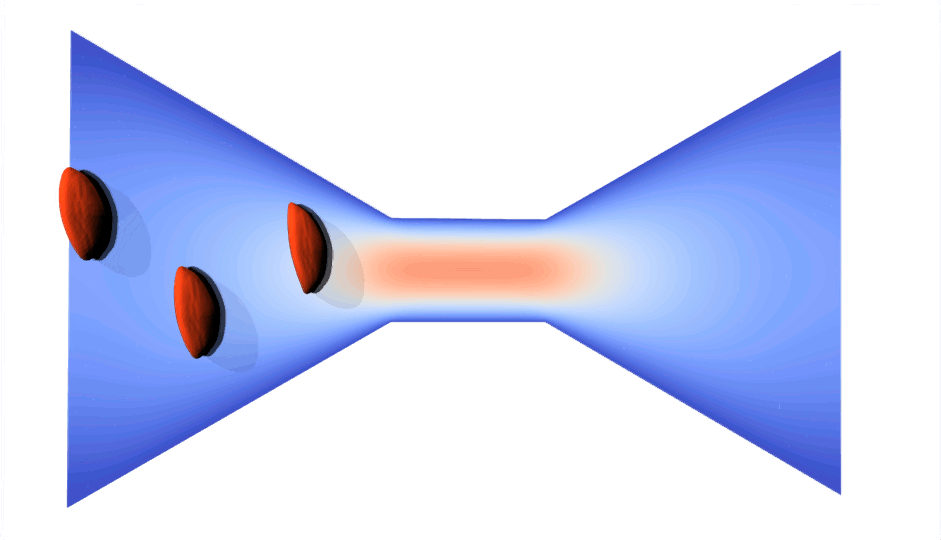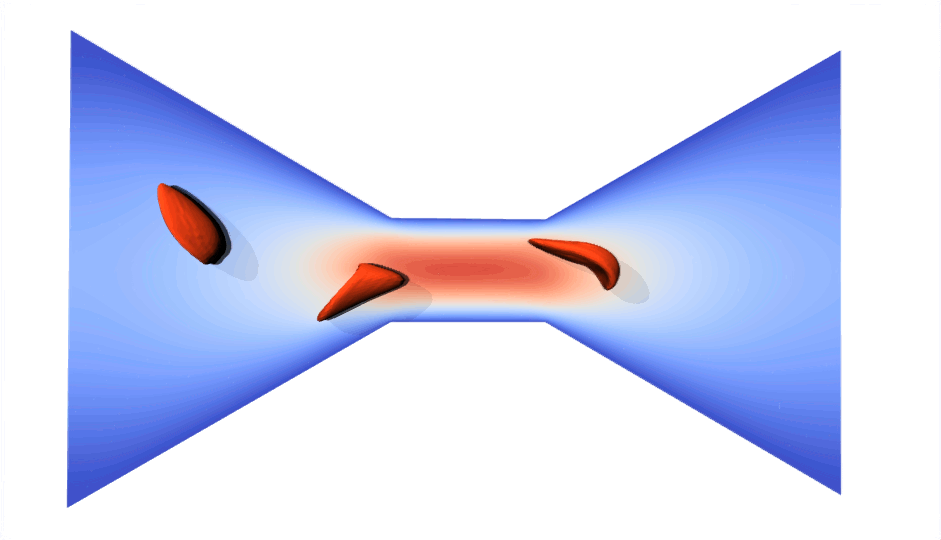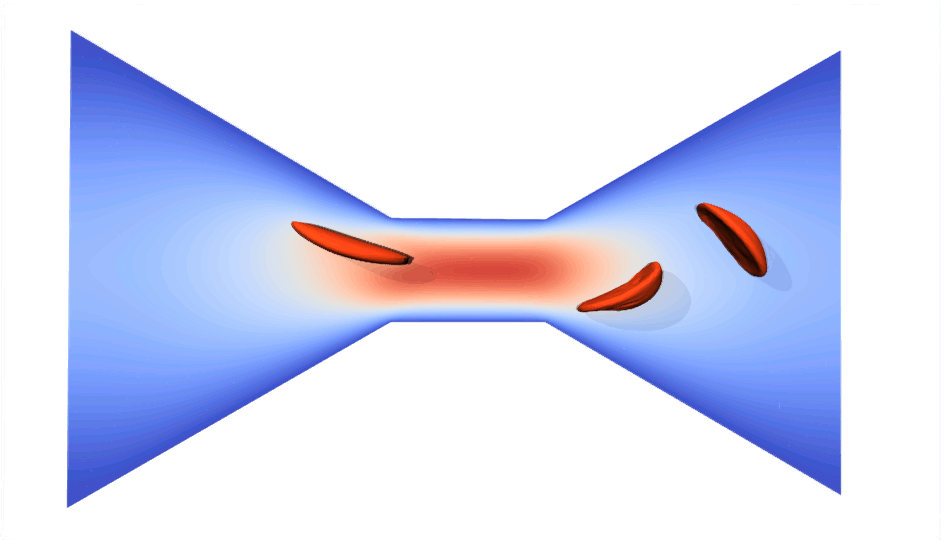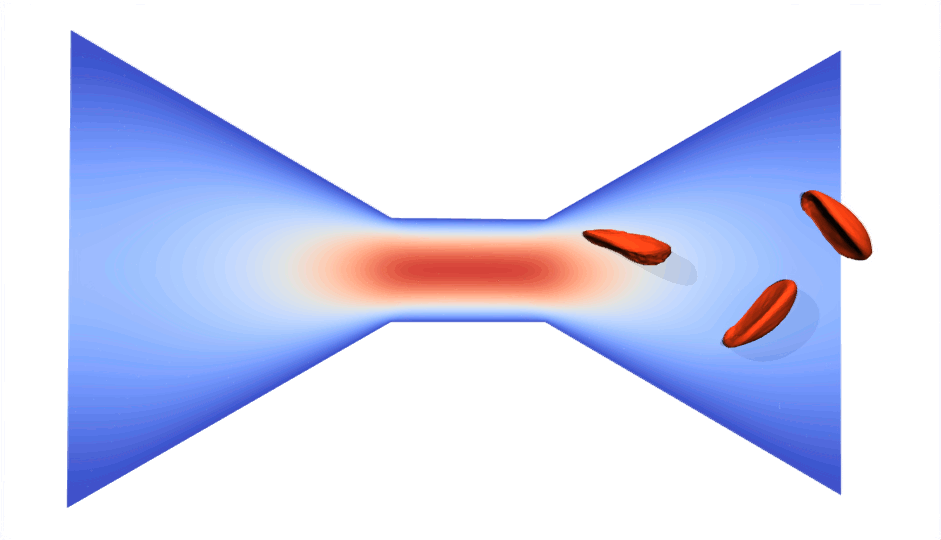
Various RBCs flowing through a microchip constriction.
You can explore the code in this github repository: PhD project